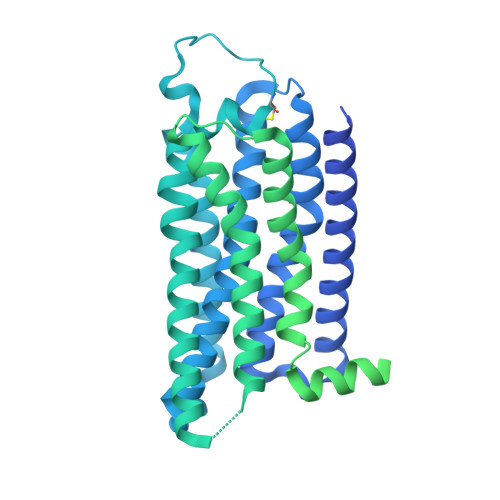
Structural insights into the agonist selectivity of the adenosine A 3 receptor.
Oshima, H.S., Ogawa, A., Sano, F.K., Akasaka, H., Kawakami, T., Iwama, A., Okamoto, H.H., Nagiri, C., Wei, F.Y., Shihoya, W., Nureki, O.(2024) Nat Commun 15: 9294-9294
- PubMed: 39511145
- DOI: https://doi.org/10.1038/s41467-024-53473-1
- Primary Citation of Related Structures:
8YH0, 8YH2, 8YH3, 8YH5, 8YH6 - PubMed Abstract:
Adenosine receptors play pivotal roles in physiological processes. Adenosine A 3 receptor (A 3 R), the most recently identified adenosine receptor, is expressed in various tissues, exhibiting important roles in neuron, heart, and immune cells, and is often overexpressed in tumors, highlighting the therapeutic potential of A 3 R-selective agents. Recently, we identified RNA-derived N 6 -methyladenosine (m 6 A) as an endogenous agonist for A 3 R, suggesting the relationship between RNA-derived modified adenosine and A 3 R. Despite extensive studies on the other adenosine receptors, the selectivity mechanism of A 3 R, especially for A 3 R-selective agonists such as m 6 A and namodenoson, remained elusive. Here, we identify tRNA-derived N 6 -isopentenyl adenosine (i 6 A) as an A 3 R-selective ligand via screening of modified nucleosides against the adenosine receptors. Like m 6 A, i 6 A is found in the human body and may be an endogenous A 3 R ligand. Our cryo-EM analyses elucidate the A 3 R-G i complexes bound to adenosine, 5'-N-ethylcarboxamidoadenosine (NECA), m 6 A, i 6 A, and namodenoson at overall resolutions of 3.27 Å (adenosine), 2.86 Å (NECA), 3.19 Å (m 6 A), 3.28 Å (i 6 A), and 3.20 Å (namodenoson), suggesting the selectivity and activation mechanism of A 3 R. We further conduct structure-guided engineering of m 6 A-insensitive A 3 R, which may aid future research targeting m 6 A and A 3 R, providing a molecular basis for future drug discovery.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan.