Cryo-EM structure of SARS-CoV-2 D614G S with one ACE2 receptor binding (RB1) in prefusion conformation
Liu, Z., Xing, L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein | A [auth C], B, C [auth A] | 1,288 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 1 Gene Names: S, 2 | 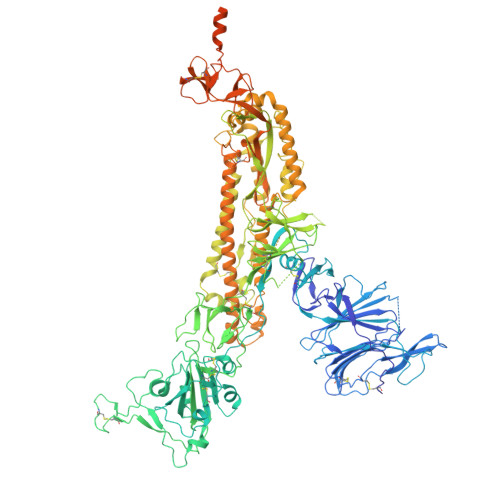 |
UniProt | |||||
Find proteins for P0DTC2 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTC2 Go to UniProtKB: P0DTC2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTC2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 16 | Go to GlyGen: P0DTC2-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Angiotensin-converting enzyme 2 | 631 | Homo sapiens | Mutation(s): 0 Gene Names: ACE2, UNQ868/PRO1885 EC: 3.4.17.23 (PDB Primary Data), 3.4.17 (PDB Primary Data) | 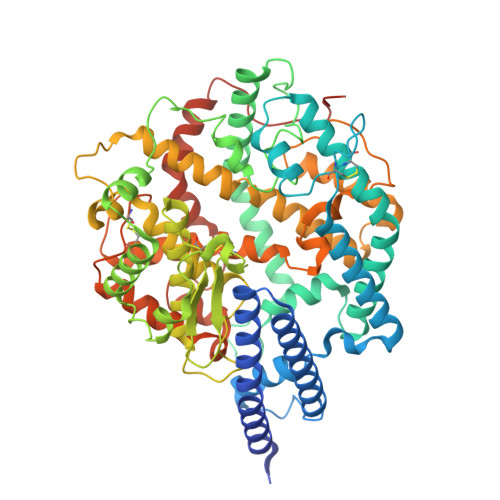 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9BYF1 (Homo sapiens) Explore Q9BYF1 Go to UniProtKB: Q9BYF1 | |||||
PHAROS: Q9BYF1 GTEx: ENSG00000130234 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9BYF1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | AA [auth C] AB [auth A] BA [auth C] BB [auth A] CA [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Science and Technology (MoST, China) | China | 2021YFC2302500 |
| National Natural Science Foundation of China (NSFC) | China | 82394463; 92169112; 82341036 |
| Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences | China | 2023-PT310-02 |
| National Key R&D Program of China | China | 2022YFC2604100; 2021YFC2300703; 2023YFC2307800 |
| Major Project of Guangzhou National Laboratory | China | GZNL2023A01008; GZNL2024A01008 |
| R&D Program of Guangzhou Laboratory | China | SRPG22-003 |
| Shanghai Municipal Science and Technology Major Project | China | ZD2021CY001 |