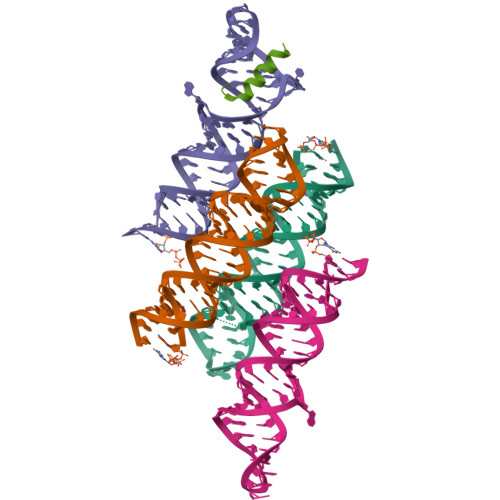
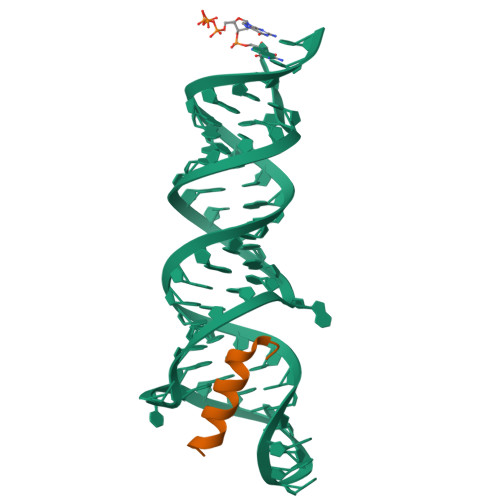
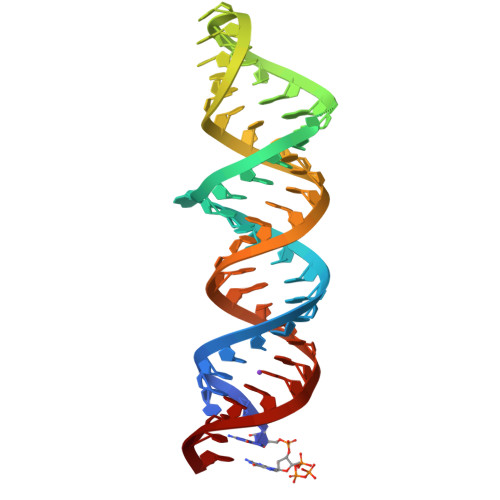
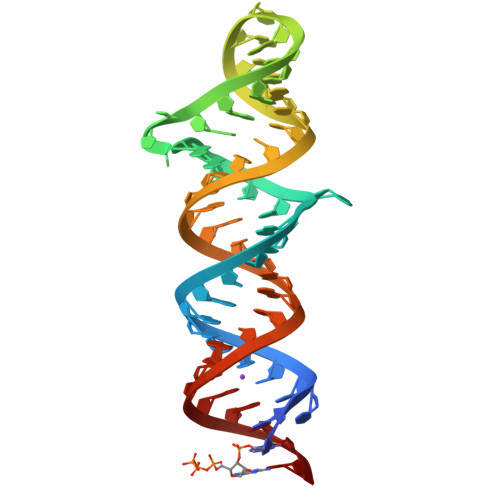
Structures of complete HIV-1 TAR RNA portray a dynamic platform poised for protein binding and structural remodeling.
Bou-Nader, C., Link, K.A., Suddala, K.C., Knutson, J.R., Zhang, J.(2025) Nat Commun 16: 2252-2252
- PubMed: 40050622
- DOI: https://doi.org/10.1038/s41467-025-57519-w
- Primary Citation of Related Structures:
9DE5, 9DE6, 9DE7, 9DE8 - PubMed Abstract:
The HIV-1 TAR RNA plays key roles in viral genome architecture, transcription and replication. Previous structural analyses focused on its upper stem loop, which has served as a paradigm to study RNA structural dynamics. However, an imperfectly paired lower stem immediately abuts and stacks with the upper half, both of which are required for efficient HIV replication. Here, we report crystal structures of the full-length HIV-1 TAR which reveal substantial conformational mobility in its three conserved bulges and in its lower stem, which coordinately maintain the structural fluidity of the entire RNA. We find that TAR RNA is a robust inhibitor of PKR, and primarily uses its lower stem to capture and sequester PKR monomers, preventing their dimerization and activation. The lower stem exhibits transient conformational excursions detected by a ligation assay. Time-resolved fluorescence spectroscopy reveals local and global TAR structural remodeling by HIV-1 nucleocapsid, Tat, and PKR. This study portrays the structure, dynamics, and interactions of a complete TAR RNA, uncovers a convergent RNA-based viral strategy to evade innate immunity, and provides avenues to develop antivirals that target a dynamic, multifunctional viral RNA.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, USA.