Inhibition of tau neuronal internalization using anti-tau single domain antibodies.
Danis, C., Dupre, E., Bouillet, T., Denechaud, M., Lefebvre, C., Nguyen, M., Mortelecque, J., Cantrelle, F.X., Rain, J.C., Hanoulle, X., Colin, M., Buee, L., Landrieu, I.(2025) Nat Commun 16: 3162-3162
- PubMed: 40175345
- DOI: https://doi.org/10.1038/s41467-025-58383-4
- Primary Citation of Related Structures:
9G13 - PubMed Abstract:
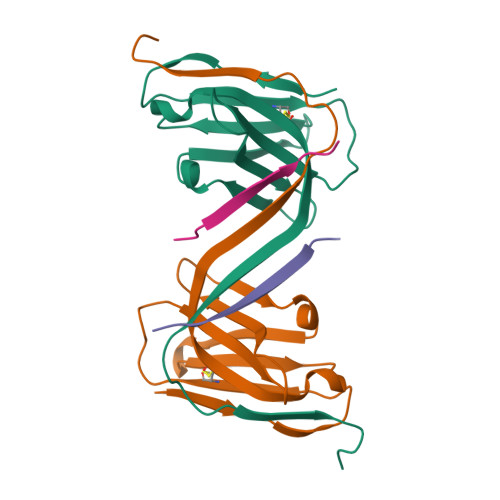
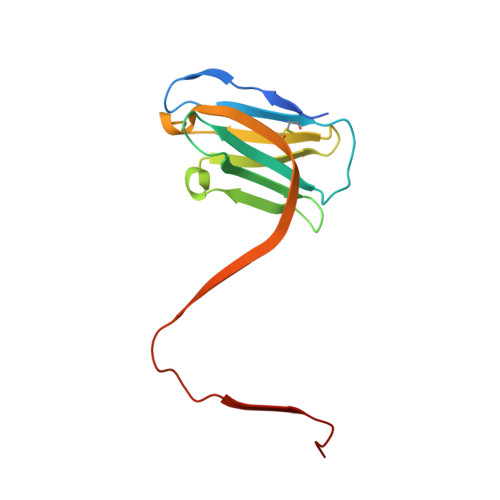
In Alzheimer's disease, tau pathology spreads across brain regions as the disease progresses. Intracellular tau can be released and taken up by nearby neurons. We evaluated single domain anti-tau antibodies, also called VHHs, as inhibitors of tau internalization. We identified three VHH inhibitors of tau uptake: A31, H3-2, and Z70 mut1 . These VHHs compete with the membrane protein LRP1, a major receptor mediating neuronal uptake of tau. A31 and Z70 mut1 bind to microtubule binding domain repeats, which are involved in the interaction with LRP1. VHH H3-2 is the only VHH from our library that reduces the internalization of both monomeric tau and tau fibrils. VHH H3-2 binds a C-terminal tau epitope with high affinity. Its three-dimensional structure in complex with a tau peptide reveals a unique binding mode as a VHH-swapped dimer. These anti-tau VHHs are interesting tools to study tau prion-like propagation in tauopathies and potentially develop novel biotherapies.
Organizational Affiliation:
CNRS EMR9002 - BSI - Integrative Structural Biology, Lille, France. clement.danis@inserm.fr.