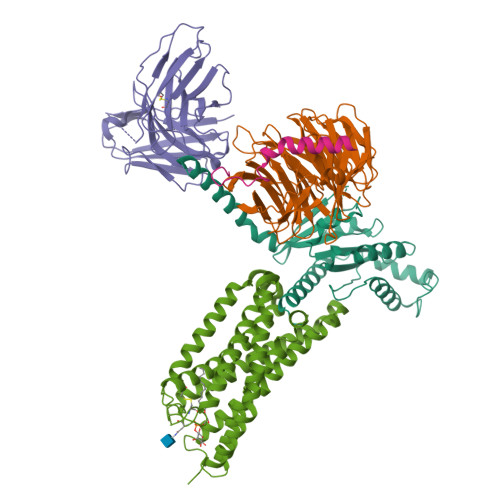
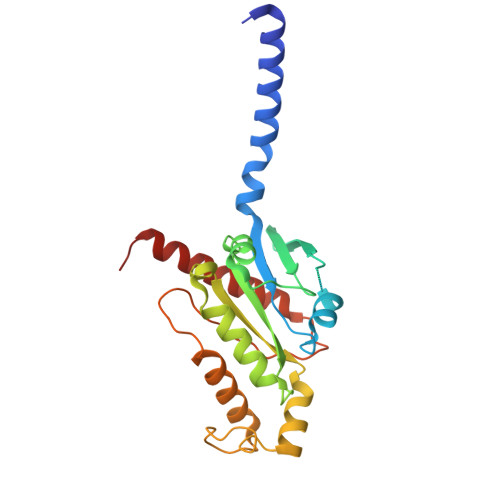
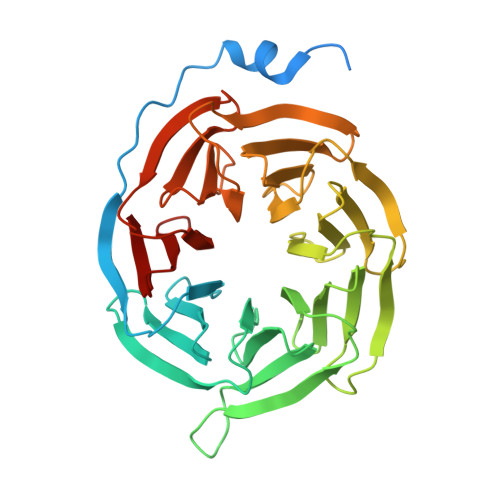
Structural basis for lipid-mediated activation of G protein-coupled receptor GPR55.
Claff, T., Ebenhoch, R., Kley, J.T., Magarkar, A., Nar, H., Weichert, D.(2025) Nat Commun 16: 1973-1973
- PubMed: 40000629
- DOI: https://doi.org/10.1038/s41467-025-57204-y
- Primary Citation of Related Structures:
9GE2, 9GE3 - PubMed Abstract:
GPR55 is an orphan G protein-coupled receptor (GPCR) and represents a promising drug target for cancer, inflammation, and metabolic diseases. The endogenous activation of lipid GPCRs can be solely mediated by membrane components and different lipids have been proposed as endogenous activators of GPR55, such as cannabinoids and lysophosphatidylinositols. Here, we determine high-resolution cryo-electron microscopy structures of the activated GPR55 in complex with heterotrimeric G 13 and two structurally diverse ligands: the putative endogenous agonist 1-palmitoyl-2-lysophosphatidylinositol (LPI) and the synthetic agonist ML184. These results reveal insights into ligand recognition at GPR55, G protein coupling and receptor activation. Notably, an orthosteric binding site opening towards the membrane is observed in both structures, enabling direct interaction of the agonists with membrane lipids. The structural observations are supported by mutagenesis and functional experiments employing G protein dissociation assays. These findings will be of importance for the structure-based development of drugs targeting GPR55.
Organizational Affiliation:
Boehringer Ingelheim Pharma GmbH & Co. KG, Global Medicinal Chemistry, Biberach an der Riß, Germany.