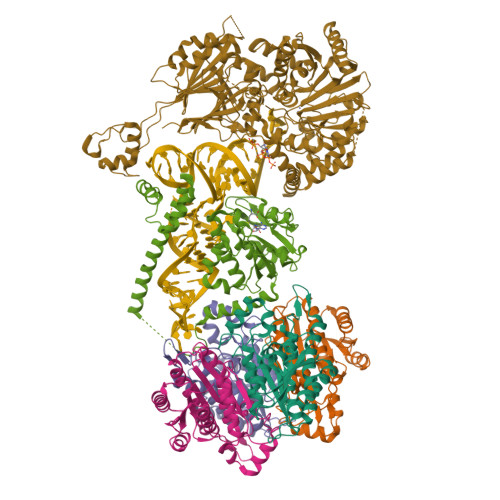
Structural basis of 3'-tRNA maturation by the human mitochondrial RNase Z complex.
Valentin Gese, G., Hallberg, B.M.(2024) EMBO J 43: 6573-6590
- PubMed: 39516281
- DOI: https://doi.org/10.1038/s44318-024-00297-w
- Primary Citation of Related Structures:
9EY0, 9EY1, 9EY2, 9GCH - PubMed Abstract:
Maturation of human mitochondrial tRNA is essential for cellular energy production, yet the underlying mechanisms remain only partially understood. Here, we present several cryo-EM structures of the mitochondrial RNase Z complex (ELAC2/SDR5C1/TRMT10C) bound to different maturation states of mitochondrial tRNA His , showing the molecular basis for tRNA-substrate selection and catalysis. Our structural insights provide a molecular rationale for the 5'-to-3' tRNA processing order in mitochondria, the 3'-CCA antideterminant effect, and the basis for sequence-independent recognition of mitochondrial tRNA substrates. Furthermore, our study links mutations in ELAC2 to clinically relevant mitochondrial diseases, offering a deeper understanding of the molecular defects contributing to these conditions.
Organizational Affiliation:
Department of Cell and Molecular Biology, Karolinska Institutet, Solna, Sweden.