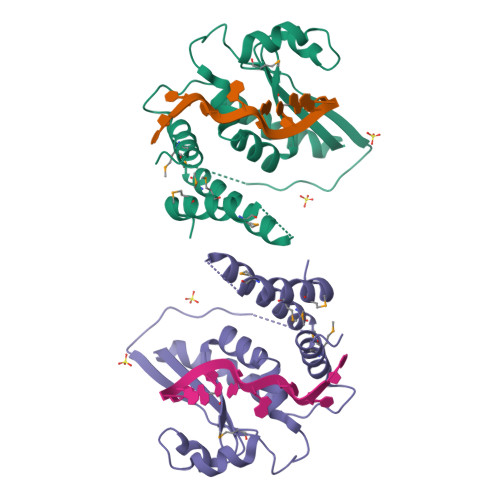
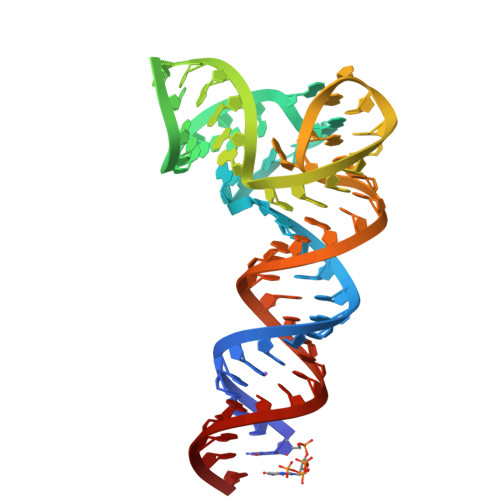
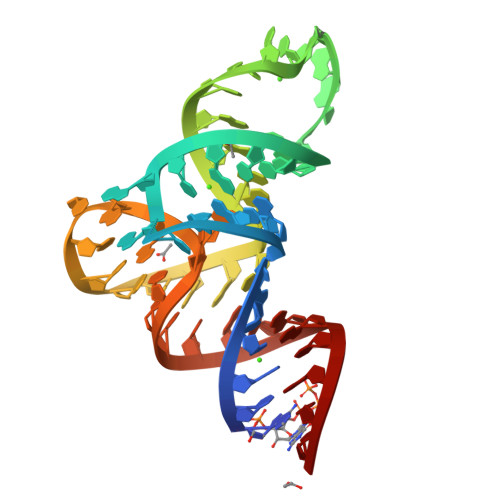
Structure Determination MethodologyScientific Name of Source OrganismRefinement Resolution (Å)Enzyme Classification Name | Blech, M. (2012) Biochem J 447: 205-215 | Released | 2012-08-22 | | Method | X-RAY DIFFRACTION 1.97 Å | | Organisms | | | Macromolecule | |
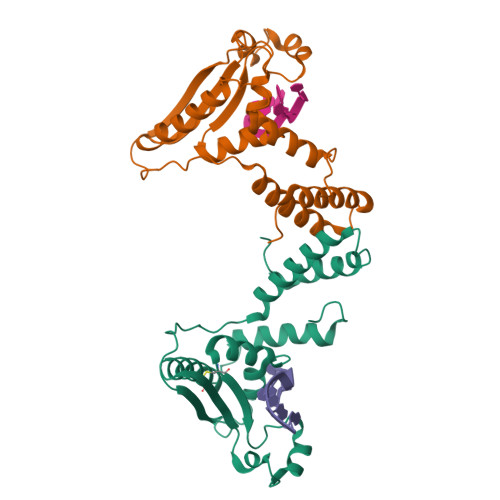
Blech, M., Hoerer, S. (2013) J Mol Biology 425: 94-111 | Released | 2012-12-19 | | Method | X-RAY DIFFRACTION 1.83 Å | | Organisms | | | Macromolecule | |
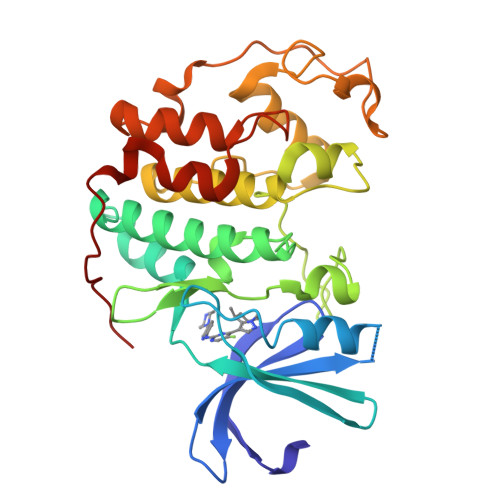
Blech, M., Hoerer, S. (2013) J Mol Biology 425: 94-111 | Released | 2012-12-19 | | Method | X-RAY DIFFRACTION 2.03 Å | | Organisms | | | Macromolecule | |
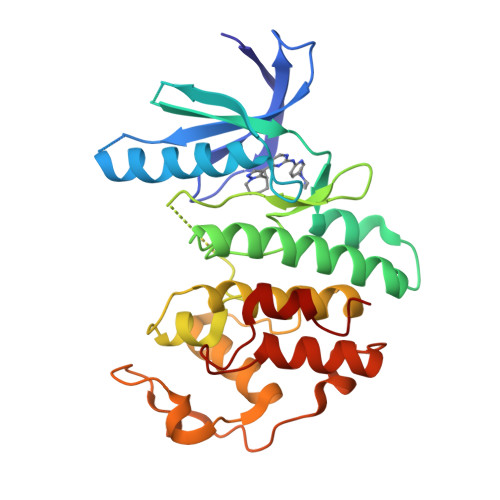
Blech, M., Hoerer, S. (2013) J Mol Biology 425: 94-111 | Released | 2012-12-19 | | Method | X-RAY DIFFRACTION 1.9 Å | | Organisms | | | Macromolecule | |
Blech, M., Hoerer, S. (2013) J Mol Biology 425: 94-111 | Released | 2012-12-19 | | Method | X-RAY DIFFRACTION 1.81 Å | | Organisms | | | Macromolecule | |
Teplova, M., Hafner, M., Teplov, D., Essig, K., Tuschl, T., Patel, D.J. (2013) Genes Dev 27: 928-940 | Released | 2013-05-08 | | Method | X-RAY DIFFRACTION 3.501 Å | | Organisms | | | Macromolecule | | | Unique Ligands | SO4 |
Teplova, M., Hafner, M., Teplov, D., Essig, K., Tuschl, T., Patel, D.J. (2013) Genes Dev 27: 928-940 | Released | 2013-05-08 | | Method | X-RAY DIFFRACTION 2.853 Å | | Organisms | | | Macromolecule | |
Murray, J.M. (2019) Bioorg Med Chem Lett 29: 2294-2301 | Released | 2020-07-29 | | Method | X-RAY DIFFRACTION 2 Å | | Organisms | | | Macromolecule | | | Unique Ligands | N14 |
Murray, J.M. (2019) Bioorg Med Chem Lett 29: 2294-2301 | Released | 2020-07-29 | | Method | X-RAY DIFFRACTION 2.707 Å | | Organisms | | | Macromolecule | | | Unique Ligands | N1A |
Murray, J.M., Boenig, G.D.L. (2019) Bioorg Med Chem Lett 29: 2294-2301 | Released | 2020-07-29 | | Method | X-RAY DIFFRACTION 1.977 Å | | Organisms | | | Macromolecule | | | Unique Ligands | N1J |
Noland, C.L., Fong, R. To be published | Released | 2023-05-03 | | Method | X-RAY DIFFRACTION 1.88 Å | | Organisms | | | Macromolecule | | | Unique Ligands | KUI |
Noland, C.L., Fong, R. To be published | Released | 2023-05-03 | | Method | X-RAY DIFFRACTION 1.78 Å | | Organisms | | | Macromolecule | | | Unique Ligands | KTF, TRS |
Valentin Gese, G., Cipullo, M., Rorbach, J., Hallberg, B.M. (2024) Nat Commun 15: 5664-5664 | Released | 2024-06-26 | | Method | ELECTRON MICROSCOPY 6.69 Å | | Organisms | | | Macromolecule | Unique protein chains: 29 Unique nucleic acid chains: 1 | | Unique Ligands | ATP, FES, GNP, K, MG, NAD, SPM, SRY, ZN |
Valentin Gese, G., Cipullo, M., Rorbach, J., Hallberg, B.M. (2024) Nat Commun 15: 5664-5664 | Released | 2024-06-26 | | Method | ELECTRON MICROSCOPY 3.34 Å | | Organisms | | | Macromolecule | Unique protein chains: 30 Unique nucleic acid chains: 1 | | Unique Ligands | ATP, FES, GDP, K, MG, NAD, SAH, ZN |
Valentin Gese, G., Cipullo, M., Rorbach, J., Hallberg, B.M. (2024) Nat Commun 15: 5664-5664 | Released | 2024-06-26 | | Method | ELECTRON MICROSCOPY 3.05 Å | | Organisms | | | Macromolecule | Unique protein chains: 31 Unique nucleic acid chains: 1 | | Unique Ligands | ATP, FES, GNP, K, MG, NAD, SPM, SRY, ZN |
Valentin Gese, G., Cipullo, M., Rorbach, J., Hallberg, B.M. (2024) Nat Commun 15: 5664-5664 | Released | 2024-06-26 | | Method | ELECTRON MICROSCOPY 2.98 Å | | Organisms | | | Macromolecule | Unique protein chains: 31 Unique nucleic acid chains: 3 | | Unique Ligands | ATP, FES, FME, GNP, GTP, K, MG, NAD, SPM, SRY, ZN |
Skeparnias, I., Zhang, J. (2024) Nat Struct Mol Biol 31: 1655-1668 | Released | 2024-07-10 | | Method | X-RAY DIFFRACTION 2.89 Å | | Organisms | | | Macromolecule | | | Unique Ligands | NA |
Skeparnias, I., Zhang, J. (2024) Nat Struct Mol Biol 31: 1655-1668 | Released | 2024-07-10 | | Method | X-RAY DIFFRACTION 2.3 Å | | Organisms | | | Macromolecule | | | Unique Ligands | ACT, CA, EDO |
|